ALSO IN THIS ISSUE
Apple Watch BP monitor gets FDA approval
Soaking up and sampling the sweat
Improving what our genomes tell us about disease risk
BEACON tracks emerging diseases as HHS shelves dashboard project
A cheat sheet of available MCEDs
The Annals of Internal Medicine journal just published (paywall) a very useful compendium of 22 multi-cancer early detection (MCED) tests with tables that summarize each test’s cancer type coverage, the methods used, and their claimed accuracy. Seven are commercially available in the US (Aristotle, EPISEEK, Galleri, OneTest, OverC MCD, Qx, and Cancerguard), although none are yet FDA-approved. Five more are available outside the US, and the remaining 10 are still in development.
COMMENTARY: We regularly evaluate MCEDs and have concluded that these tests have yet to demonstrate better health and lower mortality outcomes (although important trials are underway in the US and UK). However, for those considering taking an MCED or providing one to their patients, this paper is a great place to start. It lists which cancer types are included for each, and how accurate they claim to be.
This review makes no attempt to create an apples-to-apples accuracy comparison, a mission that’s impossible given what is currently known. Bottom line: The review shares our wait-and-see approach, stating that MCEDs are “not yet ready for prime time.” A brief video summary of their conclusions is available here.
Improving what our genomes can tell us about our disease risk
Thanks to the dramatically declining cost of genetic sequencing, it’s now potentially possible to sequence the entire genome of each newborn baby, at least in well-resourced places. (We talked about this back in January.) In theory, doing so would allow us to diagnose genetic predisposition to disease in any number of kids, thereby enabling early intervention. Two pilot programs (GUARDIAN and BeginNGS) are in the process of testing out this idea.
COMMENTARY: Well, why not? What could possibly go wrong? Next-generation sequencing (NGS) continues to become cheaper and cheaper, and for a significant minority of newborns tested, improvements in health will result. However, a recent paper in Science highlights a few of the bigger scientific problems (there are lots of ethical ones too, of course):
Except for the most obvious and serious disease-causing mutations, our knowledge of the impact of any given genetic mutation is both incomplete and unreliable. (Take a look at ClinVar, the primary repository for the clinical impact of genetic mutations, for evidence of this.)
Genetic mutations rarely do only one thing, and they rarely work alone.
Biology has ways to compensate for many genetically-driven potential problems.
Whether and how disease will result from a genetic mutation depends on clinical interventions as well as on an individual’s behavior and environmental exposures.
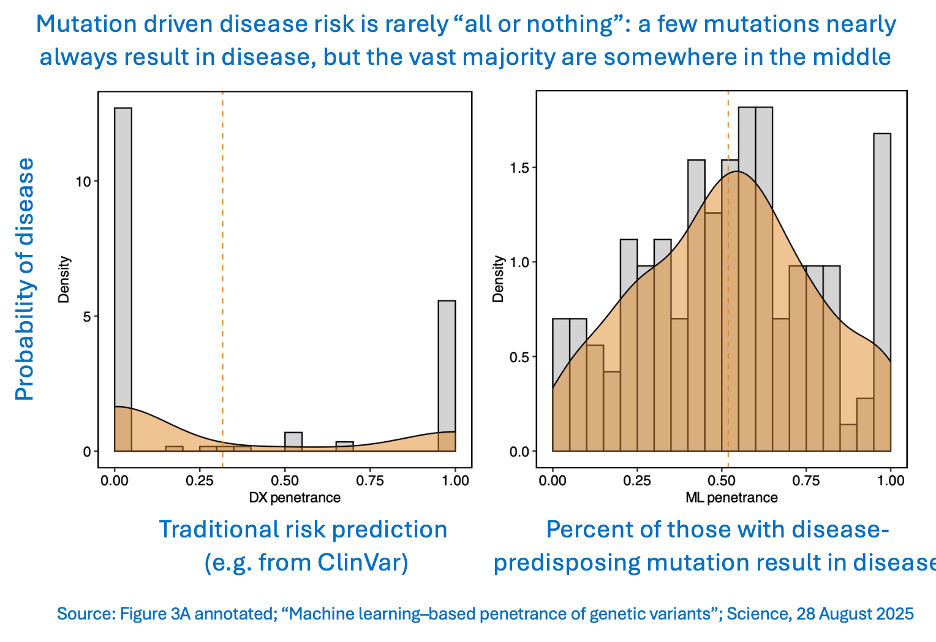
This group of issues all point to a single problem: In order to know whether a mutation is a problem or not, we need to know what its penetrance is. That’s the likelihood that someone with a harmful mutation will actually develop the associated disease. The authors used machine learning to try to solve that problem for 1,648 mutations in 31 genes known to drive any of 10 different diseases.
COMMENTARY: Traditional genetic assessment categorizes disease risk mostly into two buckets: no risk or 100% risk. But actual disease outcome is much more subtle. Only a very small subset of mutations are always pathogenic, and even fewer confer no disease risk at all.
In some ways, this is a huge study - it looked at the electronic health records of 1.3 million people for many years - but it is only a tiny excursion into the clinical impact of our inherited genome. Clearly “genes are not destiny.” But it is going to take big advances in AI’s ability to find patterns in highly complex data sets before we deeply understand exactly how genetics affect destiny. The hope is that once we do know that, we will be able to counsel patients accurately and design interventions to reduce disease penetrance in the real world.
Apple Watch BP monitor gets FDA approval
Last week we reported on the Apple Watch’s new blood-pressure-monitoring function. Only days after introducing the tool, the company announced that it had received FDA approval. Interestingly, the agency then followed up with a safety warning about “the risks related to using unauthorized devices that claim to measure or estimate blood pressure.”
Soaking up and sampling the sweat
We like sweat. (As a sample for diagnostics. Neither of us particularly enjoy sweating.) And in order to test someone’s sweat, you have to gather it - which is less straightforward than gathering blood. If you don’t care how much you get I guess you could try this nonscientific way to do it, courtesy of sci-fi cult favorite The Fifth Element. But two recent papers showcase better methods.
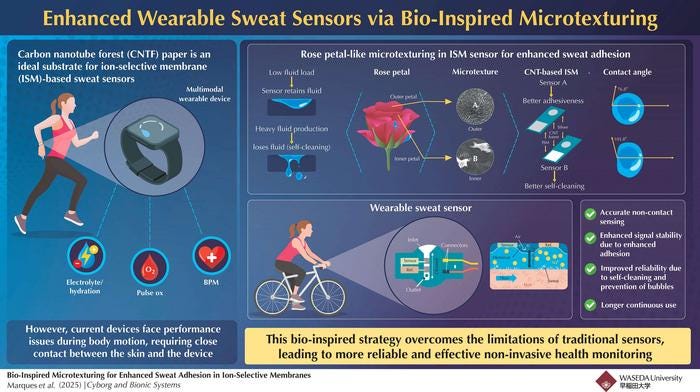
The first one aimed to address the discomfort involved with sticking a sensor directly onto the skin. They found a solution that is quite literally elegant: rose petals. When exposed to small amounts of water, their velvety surfaces pull the liquid in and allow it to stick. But when the amount of water exceeds a certain threshold, the petals suddenly repel all water, including the stuck-on droplets.
Researchers made sensors whose surface microstructure mimicked that of rose petals. Their devices were able to accurately monitor sodium concentration in sweat while leaving a 2-millimeter gap between the sensor and the skin. Plus, during periods of low sweat production, the sensors held onto fluid, and once sweat levels increased above a limit, they released the liquid. That behavior, the authors noted, keeps the signal from the sensor stable.
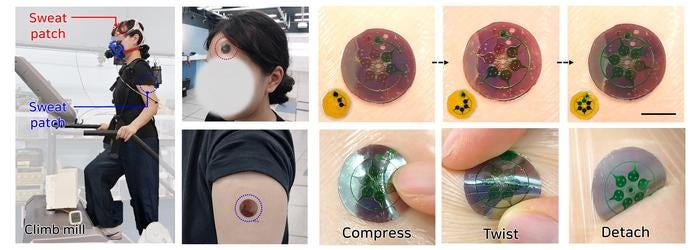
The other paper describes a sensor that also uses microchannels, but its structure achieves a different goal. It allows sweat to be collected into individual channels, one at a time (see image below). It then uses light at the nanometer scale (1/10,000th the thickness of a human hair) to analyze the sample. With this technique, the authors were able to measure how uric acid, lactic acid, and tyrosine levels changed over time.
BEACON tracks emerging diseases as HHS shelves dashboard project
Since early summer, a CDC team has been working on a dashboard that would provide real-time, searchable, state-by-state data about infectious disease in the US, MedPage Today reported last week. But just as the site was about to launch, HHS “put it on hold indefinitely.”
As the agency cuts staff and projects like its dashboard, some private organizations are stepping in to help fill the information gap left behind (though experts note that there is no way for these organizations to fully replace the scientific resources that the federal government was able to provide). This week, Boston’s NPR affiliate WBUR reported on a new university-based site that tracks emerging diseases around the world.
The Biothreats Emergence, Analysis and Communications Network (BEACON) “has posted alerts about 420 outbreaks and mapped 134 different disease-causing organisms in humans and other species” during its first three months, WBUR noted. The site is designed to be used by members of the public, not just scientists and public-health officials (though many of those folks are already taking advantage of it).
COMMENTARY: For information about what infectious diseases are doing in the US these days, we turn to two weekly newsletters: The Pandemic Center Tracking Report, from Brown University’s School of Public Health and Force of Infection, by Caitlin Rivers, epidemiologist and professor at Johns Hopkins University. We also read the news from the University of Minnesota’s Center for Infectious Disease Research and Policy (CIDRAP).
Popularized during the pandemic, wastewater can shed significant light on not only diseases in the population but what drugs that population is taking. A summer wastewater analysis for the town of Nantucket, Mass., is a case in point. The bad news: it showed cocaine levels roughly 50% above the national average. We guess the good news is that fentanyl was significantly below the national average, and methamphetamine was barely present.